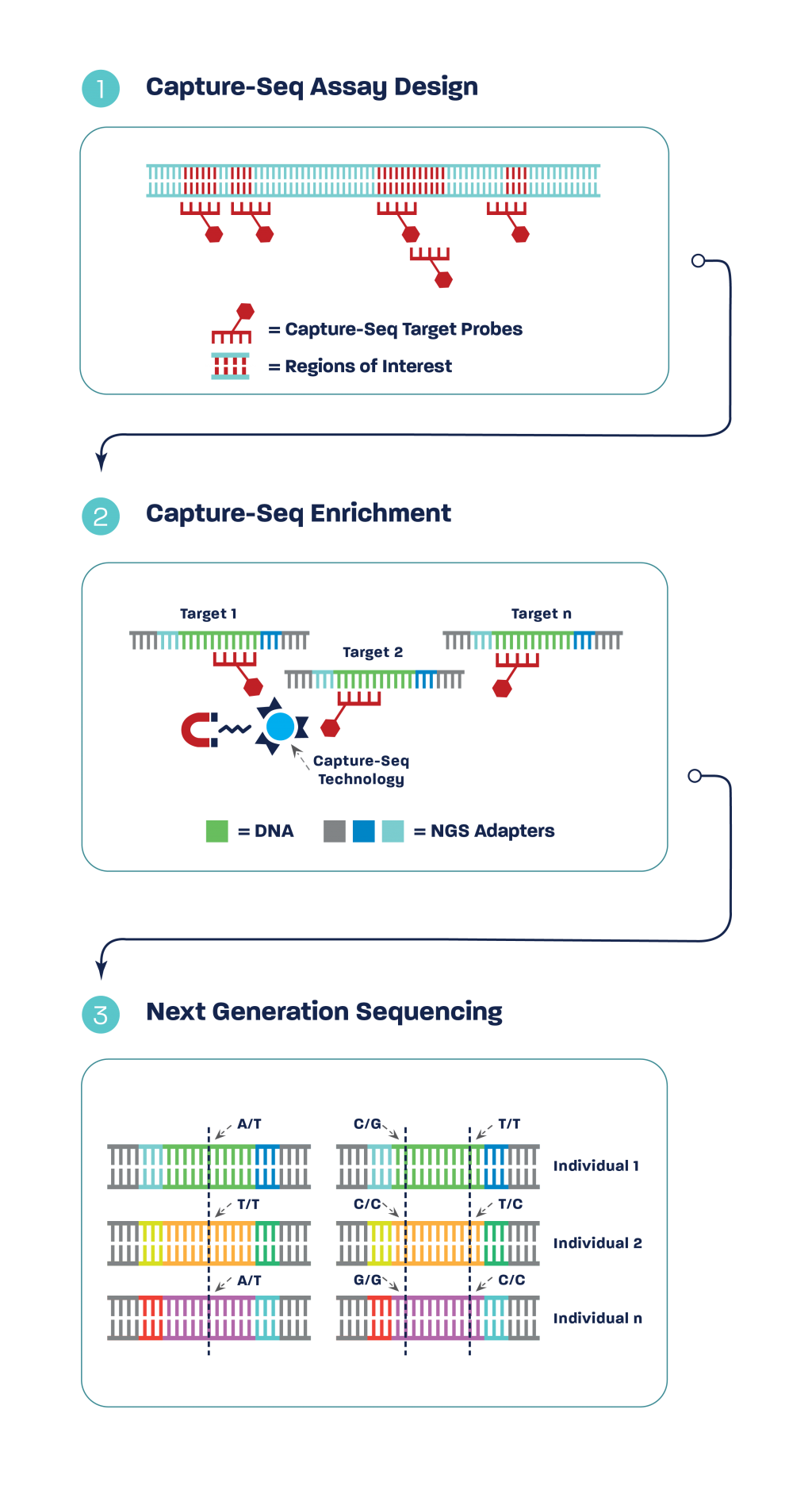
Capture-Seq is a targeted next-generation sequencing (NGS) platform for characterizing between 1,000 and 450,000+ targets in any species (including complex polyploids).
Samples may be comprised of a single species, a mix of closely related species or distantly related taxa.
Capture-Seq is suitable for both high and low volume applications. There is no minimum sample number required to utilize an existing marker assay or for custom assay design.
Capture-Seq works with complex polyploids, broad ranges of taxa, and complex host/pathogen sample types.
Capture-Seq probes can be utilized for:
| Genotyping System | Targeted Genotyping Data | Data Interval per Target | Development Cost | Ease of Customization & Adaptation | Missing Data % | Ascertainment Bias | Read Depth at Targets (Equivalent Sequencing) | Repeatability | Suitable for Novel Marker Discovery | Suitable for Haplotype REconstruction | Ease if Copy Number & Dosage Detection | Analytical Complexity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Capture-Seq | Yes | 400-500bp | Low | High | Low | Low | Predictable & Controllable | High | Yes | Yes | High | Low |
| Chip-Based Arrays | Yes | 1bp | High | Low | Low | High | N/A | High | No | No | Low | Low |
| RADSeq/GBS | No | 90bp | Low | Very Low | High | N/A | Often Low | Low | Yes | Yes | Low | High |
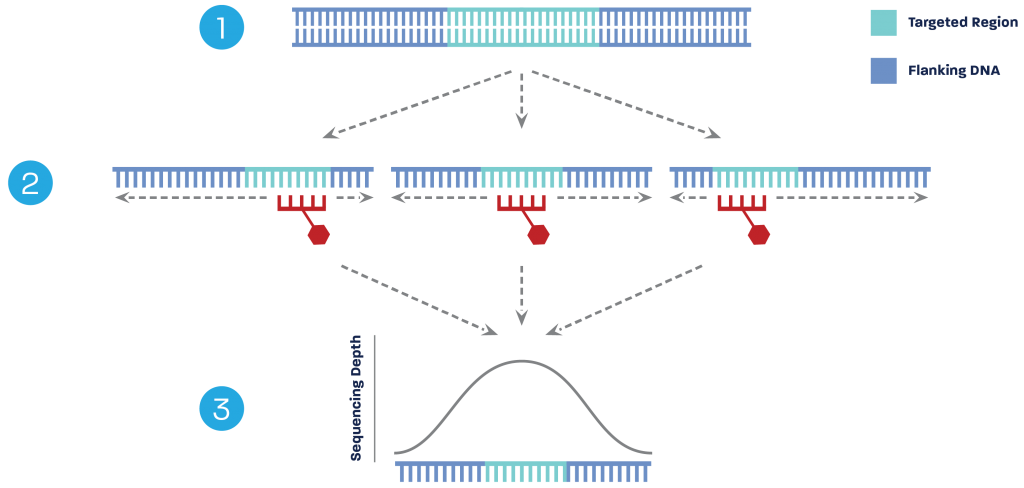
Capture-Seq delivers data both within a target region and in the adjacent regions. During sample preparation, genomic DNA is randomly fragmented (1), resulting in the target region being in the left, middle, and right of different fragments from the many genome copies within each sample.
After Capture-Seq probes hybridize to their target regions (2), during next-generation sequencing the molecules are sequenced in full. This results in sequence data obtained in the bordering / flanking regions (3).
Sequence depth is always greatest in the probe region for marker discovery and sequence characterization, with additional results discovered in the nearby regions.